
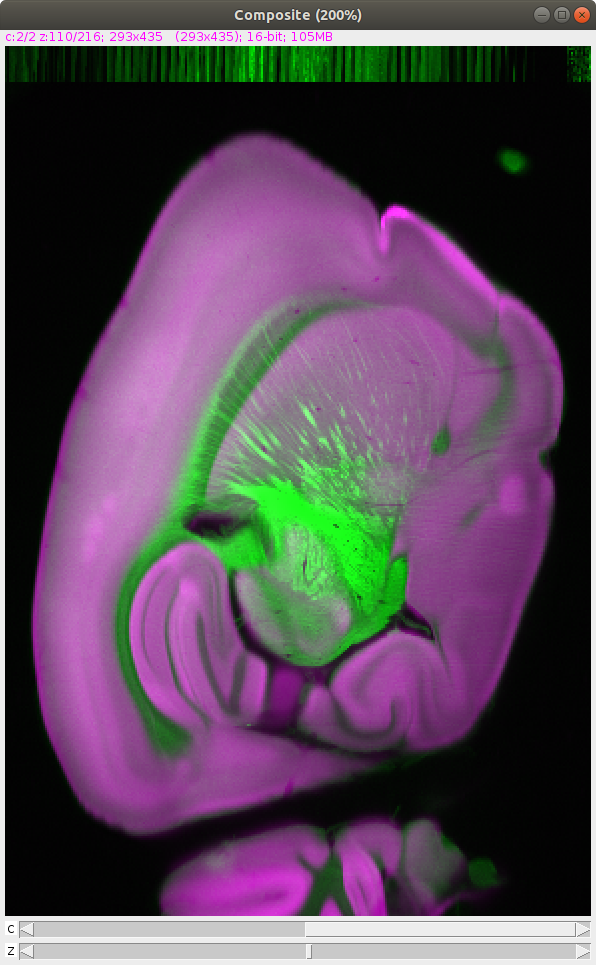
BrainJ: Whole brain reconstruction and analysis
BrainJ is a tool developed within Cellular Imaging to allow researchers to reconstruct tissue sections imaged on slide scanners and confocal microscopes into whole brains for analysis of cells, volumes, and projections. With an emphasis on accessibility and automation, this approach is easy to deploy and use, requiring no coding experience and minimal manual intervention to process multiple datasets. BrainJ is freely available on Github.
The tutorial video linked below includes a walkthrough of getting started with BrainJ, and analyze whole brain datasets, including some guides to recently added features, including:
How to automatically reconstruct and register whole brains and tissue volumes from data acquired from slide scanners and confocals.
How to automatically align these datasets to both the Allen Brain Atlas and the Enhanced and Unified Atlas (Kim et al. 2019).
How to map cells within a CCF using a variety of detection methods. Including machine learning segmentation with Ilastik.
How to perform mesoscale mapping of axons and dendrites within a CCF.
How to view and measure probe / GRIN lens placement or lesion and injection volumes within a CCF.
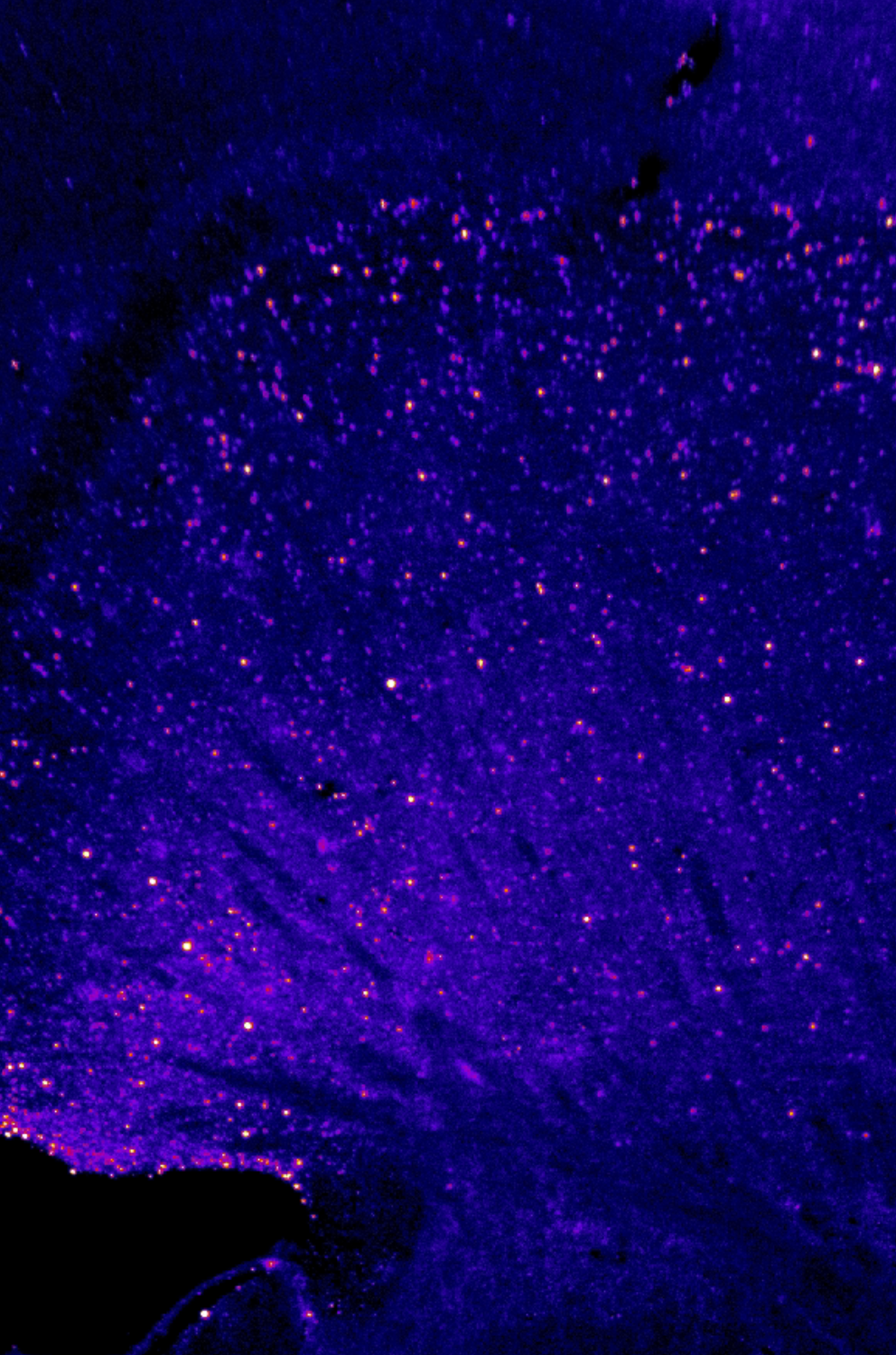
ClearMap 2: Mapping activity with CellMap
ClearMap 2 is a powerful tool for performing whole-brain analysis, but if you are new to Python, or unfamiliar with troubleshooting tools developed in Python, you will likely run into trouble getting it to work straight out of the box. This series of guides were created to help you get started, from setting up a new Linux workstation for running ClearMap, to going through how to modify the scripts so that you have a useful foundation for performing your own analysis in ClearMap.
After following our previous post, you should have a working installation of ClearMap 2 ready to analyze whole brain data.
ClearMap 2: Getting started and installation
ClearMap 2 is a powerful tool for performing whole-brain analysis, but if you are new to Python, or unfamiliar with troubleshooting tools developed in Python, you will likely run into trouble getting it to work straight out of the box. This guide is created to help you get started, from setting up a new Linux workstation for running ClearMap, to going through how to modify the scripts so that you have a useful foundation for performing your own analysis in ClearMap.
First, if you are starting a new project performing whole-brain analysis with ClearMap, it’s worth reviewing a few publications first:
Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes
Mapping the Fine-Scale Organization and Plasticity of the Brain Vasculature
The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas
The authors have also made great efforts to document both the clearing and analysis protocols, with information available at idisco.info and through existing documentation on ClearMap2.